K-Nearest Neighbors#
References#
Dataset Setup#
[5]:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.datasets import make_classification
plt.style.use('seaborn')
%matplotlib inline
[6]:
n_samples = 100
X, y = make_classification(n_samples=n_samples, n_features=2, n_redundant=0, n_classes=2, class_sep=2,
n_clusters_per_class=1, random_state=888)
X.shape, y.shape
[6]:
((100, 2), (100,))
[7]:
fig = plt.figure(figsize=(5,5))
ax1 = fig.add_subplot(1, 1, 1)
sns.scatterplot(x=X[:,0], y=X[:,1], hue=y, style=y, ax=ax1, palette='deep', s=100)
plt.show()
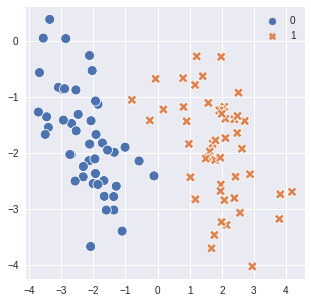
Assumption#
similar points have similar labels
Algorithm#
\begin{align*} & \text{Test Point } x\\ & \text{k nearest neighbors }S_x \subseteq D \text{, such that } | S_x | = k\\ \\ & \forall (x',y') \in D \backslash S_x \implies \text{ part of dataset but not in nearest neighbors}\\ & \text{distance } (x, x') \ge {max\atop{(x'', y'') \in S_x}} dist(x, x'') \end{align*}
In essence points that are not in k nearest neighbors than they must be farther than the test data points compared to the maximum distance from k nearest neighbors
labels of k nearest neighbors idenitfies test point’s labels.
knn is as good as the distance metric it uses.
[8]:
from scipy.spatial import minkowski_distance
[13]:
def k_nearest_neighbors(X, test_point, k):
## using minkowski distance with p2 (basically euclidean)
distances = minkowski_distance(X, test_point, p=2)
## getting closest(lowest distance) point's indeces
nearest_neighbors_indexes = np.argpartition(distances, k)[:k]
return nearest_neighbors_indexes
def plot_neighbors(X, y, test_point, ax):
sns.scatterplot(x=X[:,0], y=X[:,1], hue=y, style=y, ax=ax, palette='deep', s=100)
nearest_neighbors_indexes = k_nearest_neighbors(X, test_point, k = 7)
sns.scatterplot(x=test_point[[0]], y=test_point[[1]], color='r', ax=ax, palette='deep', marker='^',
s=150, label='test-point')
sns.scatterplot(x=X[nearest_neighbors_indexes,0], y=X[nearest_neighbors_indexes,1], color='k', ax=ax,
palette='deep', s=100, alpha=0.5, label='nearest neighbors')
ax.legend(bbox_to_anchor=(1, 1.3), loc='upper right')
learn more about distances here - https://machinelearningexploration.readthedocs.io/en/latest/MathExploration/distances.html
Training#
[14]:
fig, ax = plt.subplots(1, 3, figsize=(13, 7))
test_point1 = np.array([1.5, -1.5])
ax[0].set_title(f'test point 1 : {test_point1}')
plot_neighbors(X, y, test_point1, ax[0])
test_point2 = np.array([-2, -1])
ax[1].set_title(f'test point 2 : {test_point2}')
plot_neighbors(X, y, test_point2, ax[1])
test_point3 = np.array([0.5, -2])
ax[2].set_title(f'test point 3 : {test_point3}')
plot_neighbors(X, y, test_point3, ax[2])
plt.tight_layout()
plt.show()
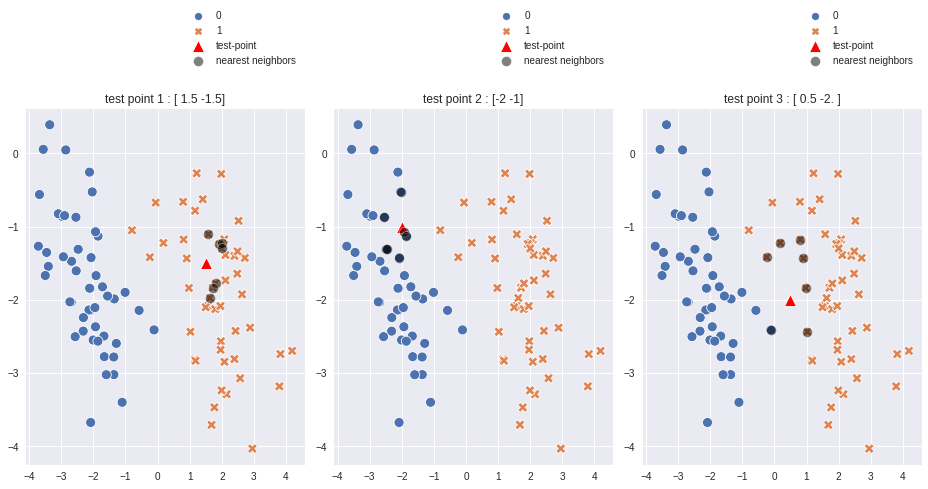
In above plot#
test point 1 = (1.5, -1.5) resides in 1’s area dominently and all 7 nearest neighbors are indicating 1 as label.
[15]:
nearest_neighbors_indexes = k_nearest_neighbors(X, test_point1, k = 7)
test_point1, y[nearest_neighbors_indexes]
[15]:
(array([ 1.5, -1.5]), array([1, 1, 1, 1, 1, 1, 1]))
test point 2 = (-2.0,-1,0) resides in 0’s area dominently and all k nearest neighbors are indicating 0 as label.
[16]:
nearest_neighbors_indexes = k_nearest_neighbors(X, test_point2, k = 7)
test_point2, y[nearest_neighbors_indexes]
[16]:
(array([-2, -1]), array([0, 0, 0, 0, 0, 0, 0]))
test point 3 = (0.5, -2) resides in somewhat center of the plot and due to euclidean distance calculation algorithm 6 out of 7 nearest neighbors have label as 1.
[17]:
nearest_neighbors_indexes = k_nearest_neighbors(X, test_point3, k = 7)
test_point3, y[nearest_neighbors_indexes]
[17]:
(array([ 0.5, -2. ]), array([1, 1, 1, 0, 1, 1, 1]))
Prediction & Decision Boundary#
[23]:
def predict(X, y, test_point, k):
nearest_neighbors_indexes = k_nearest_neighbors(X, test_point, k)
pred_y = y[nearest_neighbors_indexes]
labels, counts = np.unique(pred_y, return_counts=True)
total_possibilities = counts.sum()
pred_probs = {}
for i in zip(labels,counts):
pred_probs[i[0]] = i[1]/ total_possibilities
total_labels = np.unique(y)
for label in total_labels:
if label not in pred_probs:
pred_probs[label] = 0
return pred_probs
def prediction_boundary(X, y, k, ax):
# Creating a meshgrid to generate predictions and decision boundary.
xx, yy = np.meshgrid(np.linspace(X.min(), X.max(), 100),np.linspace(X.min() , X.max(), 100))
mesh = np.c_[xx.ravel(), yy.ravel()]
mesh_pred = np.array([predict(X, y, i, k=k).get(1) for i in mesh])
zz = mesh_pred.reshape(xx.shape)
sns.scatterplot(x=X[:,0], y=X[:,1], hue=y, style=y, ax=ax, palette='deep')
ax.contourf(xx, yy, zz, cmap=plt.cm.coolwarm, alpha=0.4)
ax.set_title(f"k = {k}")
[27]:
fig, ax = plt.subplots(1, 4, figsize=(18, 5))
prediction_boundary(X, y, k=3, ax=ax[0])
prediction_boundary(X, y, k=5, ax=ax[1])
prediction_boundary(X, y, k=7, ax=ax[2])
prediction_boundary(X, y, k=10, ax=ax[3])
plt.legend()
plt.show()
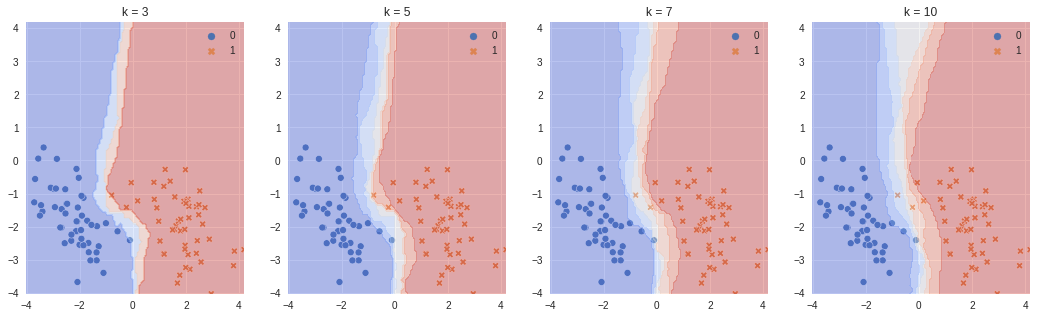
With increasing number of nearest neighbors for prediction (k) the decision boundary widens more, which means more boundary cases will have less prediction probabilties closer to 1.
